Tasks

Task 1 - Segmentation
For the segmentation task, the training data will consist of 120 T2-weighted fetal brain reconstructions from two different institutions with a corresponding label map that was manually segmented into 7 different tissues/labels, following the protocol available here:
- External Cerebrospinal Fluid
- Grey Matter
- White Matter
- Ventricles
- Cerebellum
- Deep Grey Matter
- Brainstem
The dataset consists of clinically acquired fetal brain reconstructions of both neurotypical and pathological brains, and are across a range of gestational ages.
For each case, the gestational age in weeks as well as the label neurotypical/pathological will be given in addition to the label maps. Each case will be 256x256x256 voxels, but the resolution of the cases from each institution is independent.
The testing dataset will consist of 180 cases. Included for each case is a set of label maps, consisting of the combined label map, as well as all labels separated into individual files for your convenience. The expected output from your algorithm should be the combined label map!
Task 2 - Biometry
For the biometry task, the training data will consist of biometric measurements done on the same 120 subjects included in the segmentation task above. There are five different biometry measurements, done following the annotation protocol available here:
- Brain biparietal diameter (bBIP) in the axial plane
- Skull biparietal diameter (sBIP) in the axial plane
- Height of the vermis (HV) in the sagittal plane
- Length of the corpus callosum (LCC) in the sagittal plane
- Maximum transverse cerebellar diameter (TCD) in the coronal plane.
The task will consist in predicting the measurement of the structures of interest from the super-resolution reconstructed images. The participants will be provided with a TSV file containing the target measurements for each structure, a file with the corresponding landmark in the space of the image, as well as a transform to a re-oriented space where the measurements were performed. The excepted output from your algorithm should be the length in mm of each of the structures above in a TSV or CSV file.
Note. While the ranking will be based on the the measurements, we encourage the participants to include the landmarks of their predictions if their method performs them.
Datasets
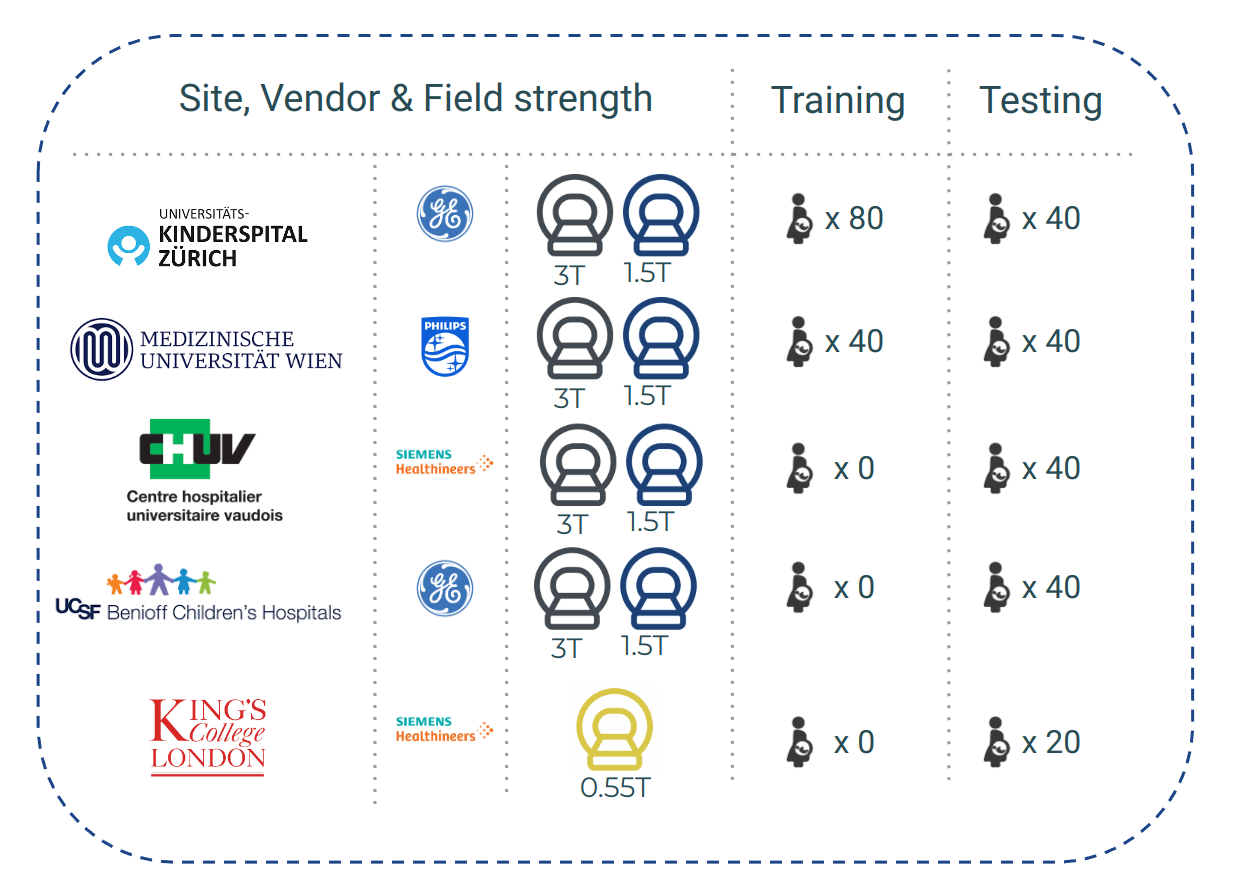
Institution 1 - University Children’s Hospital Zurich (Kispi)
Training/Testing cases: 80 / 40
Data Acquisition: The data from the University Children’s Hospital was acquired using 1.5T and 3T clinical GE whole-body scanners (Signa Discovery MR450 and MR750), either using an 8-channel cardiac coil or a body coil. T2-weighted single shot Fast Spin Echo sequences were acquired with an in-plane resolution of 0.5mm x 0.5mm, and a slice thickness of 3 to 5 mm. The sequence parameters were the following: TR: 2000-3500ms; TE: 120ms (minimum); flip angle: 90°; sampling percentages 55%. The following parameters were adjusted depending on the gestational age and size of the fetus: FOV: 200-240 mm; Image Matrix: 1.5T: 256x224; 3T: 320x224. The imaging plane was oriented relative to the fetal brain and axial, coronal, and sagittal images were acquired.
Post-Processing: For each subject, we manually reviewed the acquired fetal brain images for quality in order to compile a stack of images. Each stack consisted of at least one brain scan in each orientation, with more scans included when available. The number of scans in each stack ranged between 3 and 13. For sub-001 to sub-040, every image in the stack was then reoriented to a standard plane and a mask was created of the fetal brain using a semi-automated atlas-based custom MeVisLab (MeVis Medical Solutions AG, Bremen, Germany) module [2], [16]. An SR reconstruction algorithm was then applied to each subject’s stack of images and brain masks, creating a 3D SR volume of brain morphology (sub-001 to 040: [2]; sub-041 to sub-080: [1]) with an isotropic resolution of 0.5mm^3. Each image was histogram-matched using Slicer [8], and zero-padded to be 256x256x256 voxels.
Institution 2 - General Hospital Vienna/Medical University of Vienna
Training/Testing cases: 40 / 40
Data Acquisition: The data from the University of Vienna (training: n=40; testing: n=40) was acquired using 1.5 T (Philips Ingenia/Intera, Best, the Netherlands) and 3 T magnets (Philips Achieva, Best, the Netherlands), without the use of maternal or fetal sedation. All acquisitions were performed using a cardiac coil. For each case, at least 3 T2-weighted single-shot, fast spin echo (ssFSE) sequences (TE=80-140ms) in 3 orthogonal (axial, coronal, sagital) planes with reference to the fetal brain stem axis and/or the axis of the corpus callosum have been acquired using a 1.5 Tesla Philips Intera MR scanner. Overall, slice thickness was between 3mm and 5mm (gap 0.3-1mm), pixel spacing 0.65-1.17mm, acquisition time between 13.46 and 41.19 seconds.
Post-Processing: The preprocessing pipeline [4] consists of a data denoising step [5], followed by an in-plane super resolution [6] and automatic brain masking step [3] and concludes with a single 0.5 mm isotropic slice-wise motion correction and volumetric super-resolution reconstruction [3]. Subsequently, the resulting volumes are rigidly aligned to a common reference space [7].
Institution 3 - Lausanne University Hospital (CHUV)
Training/Testing cases: 0 / 40
Data Acquisition: The data from CHUV (testing: n=40) was acquired using 1.5 T (MAGNETOM Aera, Siemens Healthcare, Erlangen, Germany), without the use of maternal or fetal sedation. Acquisitions were performed with an 18-channel body coil and a 32-channel spine coil. Images were acquired using T2-weighted (T2W) Half-Fourier Acquisition Single-shot Turbo spin Echo (HASTE) sequences in the three orthogonal orientations; usually at least two acquisitions were performed in each orientation., TR/TE, 1200ms/90ms; flip angle, 6/23 90 ̊; echo train length, 224; echo spacing, 4.08ms; field-of-view, 360 × 360mm2 ; voxel size, 1.13 × 1.13 × 3.00mm3 ; inter-slice gap, 10%, acquisition time between 26 to 36 seconds.
Post-Processing: For each subject, the scans were manually reviewed and the good quality scans were chosen for super-resolution reconstruction, creating a 3D SR volume of brain morphology [2]. Each case was zero-padded to 256x256x256 and reoriented to a standard viewing plane.
Institution 4 - University of California San Francisco (UCSF)
Training/Testing cases: 0 / 40
Data Acquisition: The data from UCSF (tesing: n=40) was acquired using 3T GE Discovery MR750 or MR750W (wide bore) without the use of maternal or fetal sedation. Acquisitions were performed using a 32 channel GE cardiac coil. At least 3 T2-weighted ssFSE sequences were acquired with one scan per orientation (sagittal, axial, coronal) with the following parameters: 240 mm FOV with 512x512 matrix gives in plane resolution of ~0.5x0.5 mm with 3 mm slice thickness. TR is 2000-3500 ms, TE > 100 ms, 90 deg flip angle.
Post-Processing: For each subject, the scans were manually reviewed and the good quality scans were chosen for super-resolution reconstruction, creating a 3D SR volume of brain morphology [3]. Each case was zero-padded to 256x256x256 and reoriented to a standard viewing plane.
Institution 5 - King’s College London / St Thomas Hospital (KCL)
Training/Testing cases: 0 / 20
Data Acquisition: For each of the 20 cases, 6 to 9 stacks were acquired using Half-Fourier Acquisition Single-shot Turbo spin Echo (HASTE) sequence in the three orthogonal orientations with the following parameters: TR/TE, 2500ms/106ms; flip angle, 180 deg; field-of-view, 450 × 450mm2 with a 304x304 pixels base resolution; voxel size, 1.5 × 1.5 × 4.5mm3; acquisition time between 64 to 122 seconds. Acquisitions were performed using a 0.55T SIEMENS MAGNETOM Free.Max scanner (Siemens Healthineers, Erlangen, Germany) without the use of maternal or fetal sedation. All acquisitions were performed with the contour L coil and the integrated spine coil in maternal supine position
Post-Processing: For the KCL data, all available stacks were manually reviewed for quality. These with sufficient quality were automatically masked and then reconstructed to 0.8 mm isotropic resolution 3D volumes using SVRTK [9] and aligned to standard reference space and zero padded to 256x256x256.
References
[1] M. Kuklisova-Murgasova et al. “Reconstruction of fetal brain MRI with intensity matching and complete outlier removal,” Med. Image Anal., vol. 16, no. 8, pp. 1550–1564, Dec. 2012, doi: 10.1016/j.media.2012.07.004.
[2] S. Tourbier et al. “An efficient total variation algorithm for super-resolution in fetal brain MRI with adaptive regularization,” NeuroImage, vol. 118, pp. 584–597, Sep. 2015, doi: 10.1016/j.neuroimage.2015.06.018.
[3] M. Ebner et al. “An automated framework for localization, segmentation and super-resolution reconstruction of fetal brain MRI,” NeuroImage, vol. 206, p. 116324, Feb. 2020, doi: 10.1016/j.neuroimage.2019.116324.
[4] E. Schwartz et al. “The Prenatal Morphomechanic Impact of Agenesis of the Corpus Callosum on Human Brain Structure and Asymmetry,” Cereb. Cortex, vol. 31, no. 9, pp. 4024–4037, Sep. 2021, doi: 10.1093/cercor/bhab066.
[5] P. Coupe et al. “An Optimized Blockwise Nonlocal Means Denoising Filter for 3-D Magnetic Resonance Images,” IEEE Trans. Med. Imaging, vol. 27, no. 4, pp. 425–441, Apr. 2008, doi: 10.1109/TMI.2007.906087.
[6] C. Dong et al. “Accelerating the Super-Resolution Convolutional Neural Network,” in In Computer Vision - ECCV 2016, 9906 LNCS, 2016, pp. 391–407. Accessed: Dec. 13, 2021. [Online]. Available: https://www.springerprofessional.de/en/accelerating-the-super-resolution-convolutional-neural-network/10708956
[7] A. Gholipour et al. “A normative spatiotemporal MRI atlas of the fetal brain for automatic segmentation and analysis of early brain growth,” Sci. Rep., vol. 7, Jan. 2017, doi: 10.1038/s41598-017-00525-w
[8] R. Kikinis et al. “3D Slicer: A Platform for Subject-Specific Image Analysis, Visualization, and Clinical Support,” in Intraoperative Imaging and Image-Guided Therapy, Springer, New York, NY, 2014, pp. 277–289. doi: 10.1007/978-1-4614-7657-3_19.
[9] A. Uus et al. “Combined quantitative T2* map and structural T2- weighted tissue-specific analysis for fetal brain MRI: pilot automated pipeline,” PIPPI MICCAI 2023 workshop, LNCS 14246, doi: https://doi.org/10.1007/978-3-031-45544-5_3